class: center, middle, inverse, title-slide # BaseSet ## set storage and manipulation ### Lluís Revilla ### 2019/03/29 (updated: 2019-03-28) --- # Where do I started? A set is a group of elements, so we can define elements and the name they are grouped: 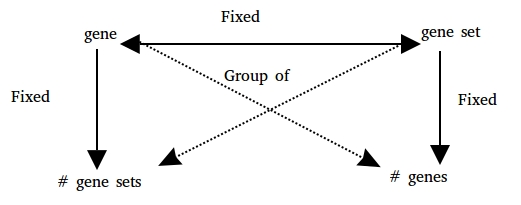 Testing other methods from this prespective in [GSEAdv](https://github.com/llrs/GSEAdv) but it took to long for some methods, that's when I started exploring with BaseSet ```r library("BaseSet") ``` Later on I found that there was a discussion about making GSEABase faster/more efficient --- ## Sets - With a data.frame: ```r relations <- data.frame(sets = c(rep("a", 5), "b"), elements = letters[seq_len(6)]) (TS1 <- BaseSet::tidySet(relations)) ## elements sets ## 1 a a ## 2 b a ## 3 c a ## 4 d a ## 5 e a ## 6 f b ``` - With a list: ```r relations <- list(a = letters[seq_len(5)], b = letters[6]) (TS2 <- BaseSet::tidySet(relations)) ## elements sets ## 1 a a ## 2 b a ## 3 c a ## 4 d a ## 5 e a ## 6 f b ``` --- # Integrating with GSEABase Converting from previous data formats ## From GeneSet ```r gs1 <- GeneSet(setName = "set1", setIdentifier = "101", geneIds = c("a", "b")) gs2 <- GeneSet(setName = "set2", setIdentifier = "102", geneIds = c("b", "c")) (GS1 <- BaseSet::tidy(gs1)) ## elements sets Identifier shortDescripton longDescription organism ## 1 a set1 101 ## 2 b set1 101 (GS2 <- BaseSet::tidy(gs2)) ## elements sets Identifier shortDescripton longDescription organism ## 1 b set2 102 ## 2 c set2 102 ``` --- ## From GeneSetCollection ```r gsc <- GeneSetCollection(gs1, gs2) (TS_gsc <- BaseSet::tidy(gsc)) ## elements sets Identifier shortDescripton longDescription organism ## 1 a set1 101 ## 2 b set1 101 ## 3 b set2 102 ## 4 c set2 102 ``` --- ## From GeneColorSet ```r data("sample.ExpressionSet", package = "Biobase") gcs1 <- GeneColorSet(sample.ExpressionSet[100:109], phenotype = "imaginary") TS <- BaseSet::tidy(gcs1) BaseSet::sets(TS) ## sets Identifier ## 1 set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## shortDescripton ## 1 Smoking-Cancer Experiment ## longDescription organism ## 1 An example object of expression set (ExpressionSet) class Homo sapiens ## pubMedIds urls contributor type ## 1 www.lab.not.exist Pierre Fermat ExpressionSet BaseSet::elements(TS) ## elements ## 1 31339_at ## 2 31340_at ## 3 31341_at ## 4 31342_at ## 5 31343_at ## 6 31344_at ## 7 31345_at ## 8 31346_at ## 9 31347_at ## 10 31348_at ``` --- ```r TS ## elements sets Identifier ## 1 31339_at set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## 2 31340_at set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## 3 31341_at set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## 4 31342_at set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## 5 31343_at set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## 6 31344_at set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## 7 31345_at set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## 8 31346_at set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## 9 31347_at set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## 10 31348_at set1 IBP10001:16652:Thu Mar 28 16:57:37 2019:20 ## shortDescripton ## 1 Smoking-Cancer Experiment ## 2 Smoking-Cancer Experiment ## 3 Smoking-Cancer Experiment ## 4 Smoking-Cancer Experiment ## 5 Smoking-Cancer Experiment ## 6 Smoking-Cancer Experiment ## 7 Smoking-Cancer Experiment ## 8 Smoking-Cancer Experiment ## 9 Smoking-Cancer Experiment ## 10 Smoking-Cancer Experiment ## longDescription organism ## 1 An example object of expression set (ExpressionSet) class Homo sapiens ## 2 An example object of expression set (ExpressionSet) class Homo sapiens ## 3 An example object of expression set (ExpressionSet) class Homo sapiens ## 4 An example object of expression set (ExpressionSet) class Homo sapiens ## 5 An example object of expression set (ExpressionSet) class Homo sapiens ## 6 An example object of expression set (ExpressionSet) class Homo sapiens ## 7 An example object of expression set (ExpressionSet) class Homo sapiens ## 8 An example object of expression set (ExpressionSet) class Homo sapiens ## 9 An example object of expression set (ExpressionSet) class Homo sapiens ## 10 An example object of expression set (ExpressionSet) class Homo sapiens ## pubMedIds urls contributor type ## 1 www.lab.not.exist Pierre Fermat ExpressionSet ## 2 www.lab.not.exist Pierre Fermat ExpressionSet ## 3 www.lab.not.exist Pierre Fermat ExpressionSet ## 4 www.lab.not.exist Pierre Fermat ExpressionSet ## 5 www.lab.not.exist Pierre Fermat ExpressionSet ## 6 www.lab.not.exist Pierre Fermat ExpressionSet ## 7 www.lab.not.exist Pierre Fermat ExpressionSet ## 8 www.lab.not.exist Pierre Fermat ExpressionSet ## 9 www.lab.not.exist Pierre Fermat ExpressionSet ## 10 www.lab.not.exist Pierre Fermat ExpressionSet ``` --- ## Operations Convert ids: ```r library("magrittr") library("hgu95av2.db") BaseSet::mutate_element(TS, symbol = mapIds(hgu95av2.db, keys = as.character(elements), keytype = "PROBEID", column = "SYMBOL"), elements = mapIds(hgu95av2.db, keys = as.character(elements), keytype = "PROBEID", column = "ENTREZID")) %>% BaseSet::elements() ## 'select()' returned 1:1 mapping between keys and columns ## 'select()' returned 1:1 mapping between keys and columns ## elements symbol ## 1 51050 PI15 ## 2 9313 MMP20 ## 3 3748 KCNC3 ## 4 2590 GALNT2 ## 5 3557 IL1RN ## 6 28778 IGLV6-57 ## 7 9407 TMPRSS11D ## 8 26584 DUX1 ## 9 3537 IGLC1 ## 10 <NA> <NA> ``` --- - Intersection ```r TS_gsc %>% BaseSet::intersection(c("set1", "set2")) ## elements sets Identifier shortDescripton longDescription organism ## 1 b set1∩set2 <NA> <NA> <NA> <NA> ``` - Union ```r BaseSet::union(TS_gsc, c("set1", "set2")) ## Error: Duplicated sets but with different information sets(TS_gsc) <- sets(TS_gsc)[, 1, drop = FALSE] BaseSet::union(TS_gsc, c("set1", "set2")) ## elements sets ## 1 a set1∪set2 ## 2 b set1∪set2 ## 3 c set1∪set2 ``` - Incidence ```r BaseSet::incidence(TS_gsc) ## set1 set2 ## a 1 0 ## b 1 1 ## c 0 1 ``` --- ## Fuzzy sets When doing several analysis different results or bootstrapping on the same result it might be important to store the strength of this relationship: - With a data.frame: ```r relations <- data.frame(sets = c(rep("a", 5), "b"), elements = letters[seq_len(6)], fuzzy = runif(6)) (TS1 <- BaseSet::tidySet(relations)) ## elements sets fuzzy ## 1 a a 0.73890495 ## 2 b a 0.10801634 ## 3 c a 0.89345813 ## 4 d a 0.12541371 ## 5 e a 0.72362393 ## 6 f b 0.01386686 ``` --- - With a list: ```r A <- runif(5) names(A) <- letters[seq_len(5)] B <- runif(1) names(B) <- letters[3] relations <- list(A = A, B = B) (TS2 <- BaseSet::tidySet(relations)) ## elements sets fuzzy ## 1 a A 0.89259590 ## 2 b A 0.14489438 ## 3 c A 0.44805864 ## 4 c B 0.05329086 ## 5 d A 0.17113448 ## 6 e A 0.43514494 ``` --- - Intersection ```r BaseSet::intersection(TS2, c("A", "B")) ## elements sets fuzzy ## 1 c A∩B 0.05329086 ``` - Union ```r BaseSet::union(TS2, c("A", "B")) ## elements sets fuzzy ## 1 a A∪B 0.8925959 ## 2 b A∪B 0.1448944 ## 3 c A∪B 0.4480586 ## 4 d A∪B 0.1711345 ## 5 e A∪B 0.4351449 ``` - Incidence ```r BaseSet::incidence(TS2) ## A B ## a 0.8925959 0.00000000 ## b 0.1448944 0.00000000 ## c 0.4480586 0.05329086 ## d 0.1711345 0.00000000 ## e 0.4351449 0.00000000 ```